Welcome to 2021 everyone!
It is with pleasure that we announce the availability of searchable PDFs of the thesis manuscript of A.B. de Haan, and the thesis of Gerhard Asmussen.
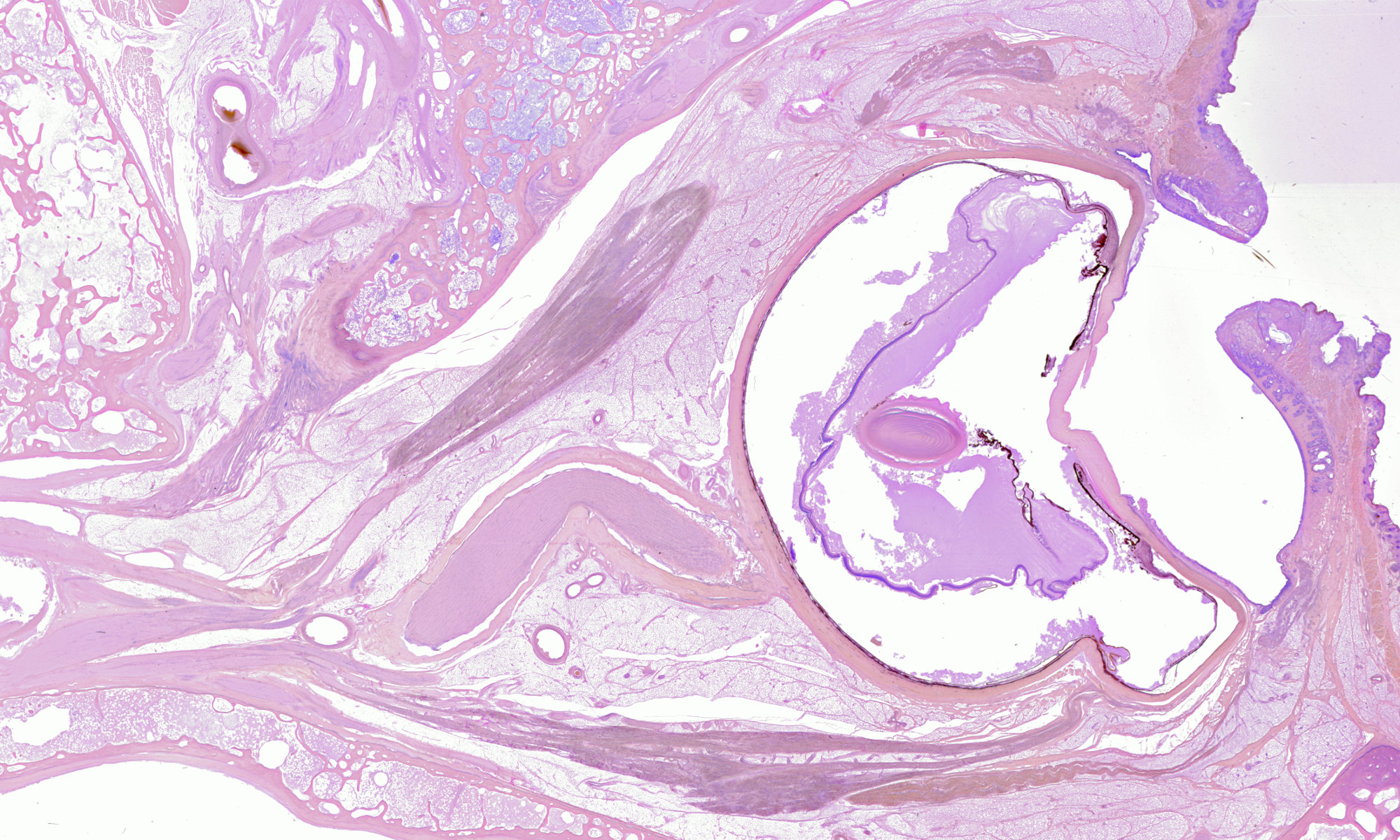
Digital 3-D reconstructions of orbital anatomy
Welcome to 2021 everyone!
It is with pleasure that we announce the availability of searchable PDFs of the thesis manuscript of A.B. de Haan, and the thesis of Gerhard Asmussen.
It’s almost exactly 13 years since the very first entry on this site, the Visible Orbit blog.
I’ve just moved the whole site, including 50GB of high resolution orbita and embryo histology slices, to its new home on a server in Nuremburg.
Everything should feel slightly snappier and a little more fresh, but most importantly: All of the resources here are still available at the same locations for 13 years now, and things are looking good for the coming years.
On our high-res section stacks page, you can now find seven fully registered stacks of our highest resolution scans of 4 orbits, 2 embryonic heads and 1 full embryo. All of the 1500 slices in these sets have been carefully registered (mapped, in this case by rotation and translation) onto each other using the fantastic TrakEM2 plugin of the Fiji ImageJ distribution.
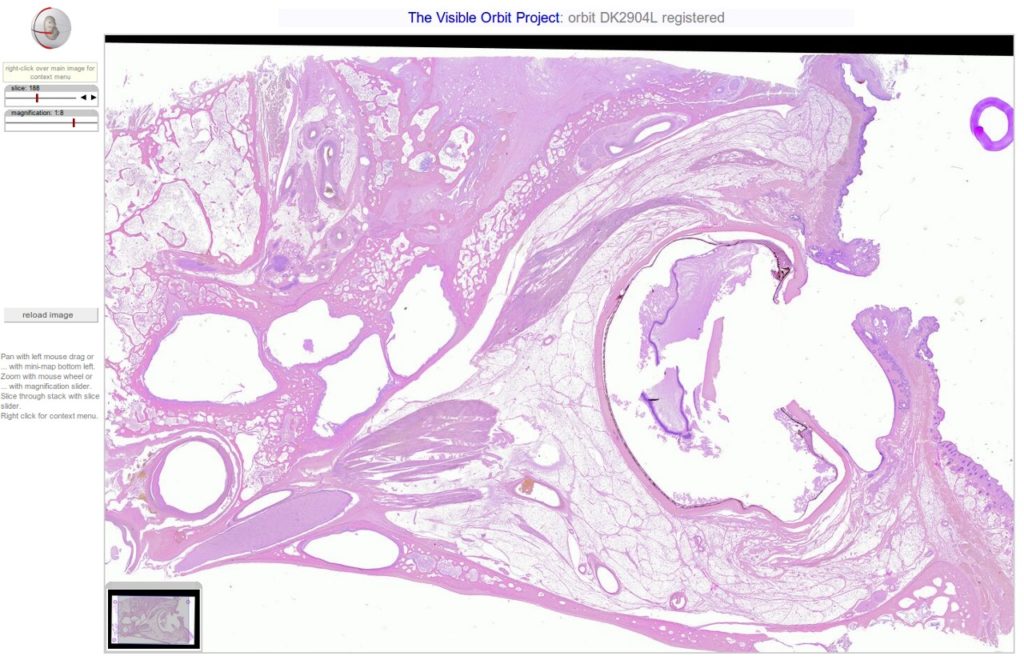
Were very happy to announce that tonight weve posted 5 new orbit section stacks totalling 1724 high resolution slices. To view the stacks using the Woolz-IIP viewer, go to our high-res section stacks page!
Today this server was upgraded: Double the RAM, lots more disc space and RHEL 6.1. This also meant that we could finally upgrade to WordPress 3.2.1, and whilst we were at it we installed a brand new theme and tweaked the PHP setup for more speed.
With some help from the Edinburgh Mouse Atlas Project (EMAP), we also now have the Woolz IIP server running on an internal test machine on one of the orbita slice stacks. At the moment, more of our machines are crunching all off the stacks into pyramidal tiffs, and then well start moving theme here where they belong. This means youll be able to browse through all the stacks at full resolution, Google Maps style!
The source code to HistoVis, the client-server visualisation framework developed by Jeroen Verschuur, has been available online for some time now. However, between the technical challenges of getting it going on the aging Redhat installation on the Sun server at our disposal and general time-pressure, the system is not operational on the visible orbit data yet.
For this and other reasons, we have recently decided to make the high resolution slices available here via the Woolz IIP viewer software as soon as possible. Please bear with us as we get the gigabytes of data pre-processed and ready. At such time, you’ll be able to view any of the high-resolution slices in your browser. For an example, see the TS19 stack on the Woolz IIP viewer website.
At the end of last year, Jeroen Verschuur successfully defended his M.Sc. thesis on the development of a remote client-server visualisation framework for large histological datasets.
We are currently in the process of getting his source code ready for an open source release. At the same time, we are planning to roll out his system, including 3D viewer client, on this server, initially with a subset of the full orbita data collection. Over time, we’ll continue adding more datasets to the viewer.
We’ll keep you posted on our progress!
Two of our project members, Prof. Huib Simonsz and Sander Schutte, are at the ARVO 2008 in Florida. Sander will be presenting a talk “Biomechanics of Orbital Fat and Eye Muscles” on Sunday morning at 10:00 in the Grand Floridian A/B.
On Thursday he will be presenting the Visible Orbit poster “A Client-Server System for Remote 3D-Visualization of Orbital Anatomy” in Hall B/C. The poster details our current progress on the new client-server 3-D visualization system for histological data. Be sure to stop by and say hello!
Stay tuned for more site updates in the following days.
By the time you read this, this website will have gone live. Going to http://visible-orbit.org/ or to http://visibleorbit.org/ will send you right here.
I’ve ported and updated most of the content from the old website. Sincere thanks to Sim IJskes and QCG for hosting us all this time.
The new Visible Orbit site is coming to you from a brand-new RHEL-powered Sun server hosted at the TU Delft data centre. Please bear with us as we port content from the old site.